Why is circular RNA (circRNA) so hot? What's it all about?
CircRNA, or circular RNA, is a class of non-coding RNA molecules that have a closed, circular structure. They lack 5' caps and 3' poly(A) tails, and are mainly found in the cytoplasm or stored in extracellular vesicles. CircRNAs are relatively stable and are not easily degraded by RNA exonucleases. They are widely present in a variety of eukaryotic organisms.
Classification
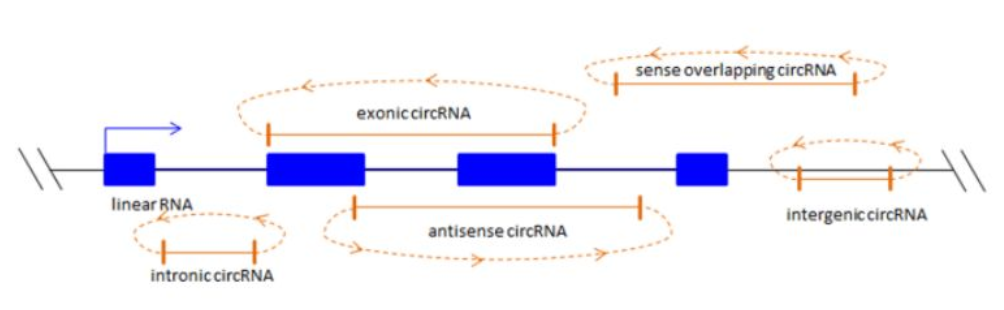
CircRNAs can be classified according to their origin of formation.
Exonic circRNAs: These are formed by the internal re-ligation of exons during RNA splicing.
Intronic circRNAs: These are formed by the internal re-ligation of introns during RNA splicing.
Lariat circRNAs: These are formed by the internal re-ligation of an exon and an intron, with the exon forming the loop of the lariat.
CircRNAs derived from viral RNA genomes, tRNA, rRNA, snRNA, etc.: These are formed by the internal re-ligation of RNA molecules from these sources.
Biological functions
CircRNAs are involved in a wide range of biological processes, including:
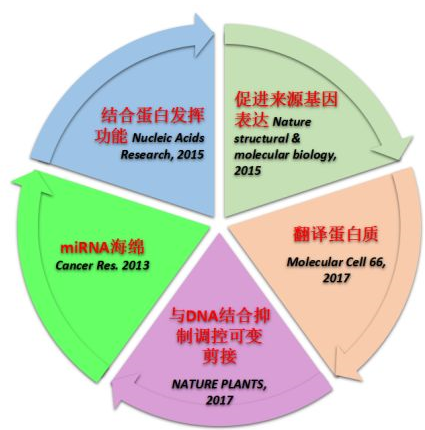
Regulation of gene expression: CircRNAs can regulate gene expression by a variety of mechanisms, including:
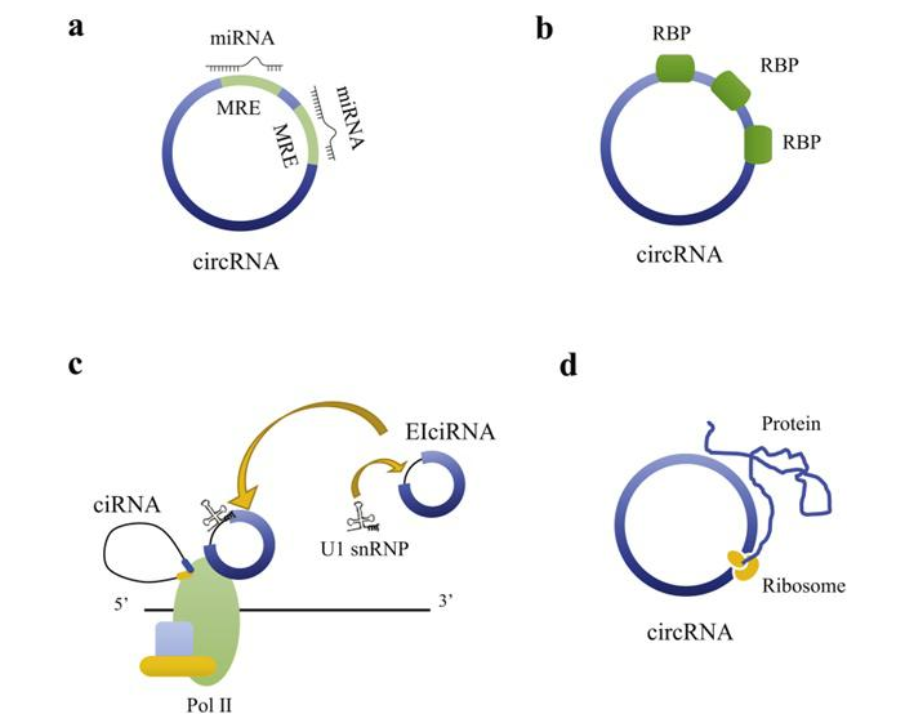
Acting as miRNA sponges: CircRNAs can bind to miRNAs and prevent them from binding to their target mRNAs, thereby suppressing gene expression.
Binding to RNA-binding proteins (RBPs): CircRNAs can bind to RBPs and alter the stability or splicing of target mRNAs.
Directly binding to transcription factors: CircRNAs can bind to transcription factors and regulate gene transcription.
Cellular signaling: CircRNAs can participate in cellular signaling by regulating the activity of signaling pathways.
Development and differentiation: CircRNAs play important roles in development and differentiation.
Disease: CircRNAs are involved in the pathogenesis of a variety of diseases, including cancer, neurodegenerative diseases, and autoimmune diseases.
Applications
CircRNAs have a number of potential applications, including:
Diagnosis and prognosis of diseases: CircRNAs can be used as biomarkers for the diagnosis and prognosis of diseases.
Targeted therapy: CircRNAs can be targeted for therapeutic intervention.
Drug discovery: CircRNAs can be used to identify new drug targets.
Technical process
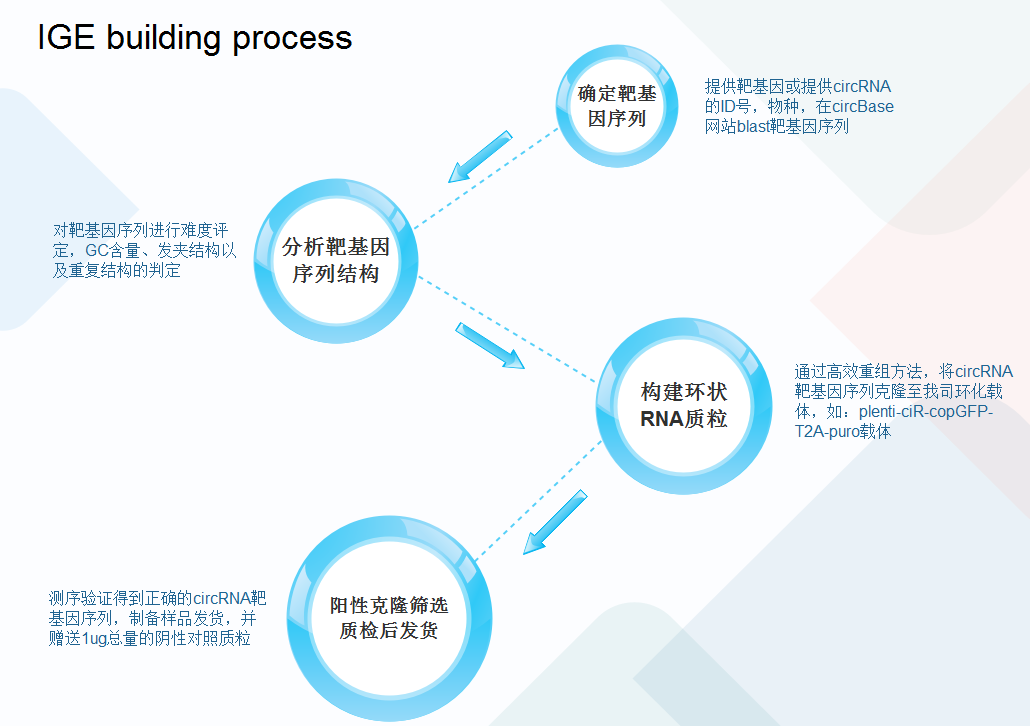
Service advantages
1.High circularization efficiency and stability
IGEBio's proprietary upstream and downstream circularization frameworks are well-suited for circularizing target sequences of various lengths. All validated circRNAs have a circularization efficiency of at least 50%, which can effectively ensure at least 100-fold overexpression efficiency.
2.Convenient service
IGEBio provides a prediction service for circRNA coding ability. Users only need to provide the circRNA ID and species to order the construction service. A negative control plasmid with a total amount of 1 ug is provided free of charge.
3.Accurate sequence circularization
The specific circularization-mediated sequence introduced into the circularization framework can effectively guarantee the accurate circularization of the linear sequence of the target circRNAs.
4.High system diversity
IGEBio can provide cell-based transient and lentiviral expression systems. It can also provide systems that express GFP fluorescence or resistance genes such as puromycin, which is convenient for screening.
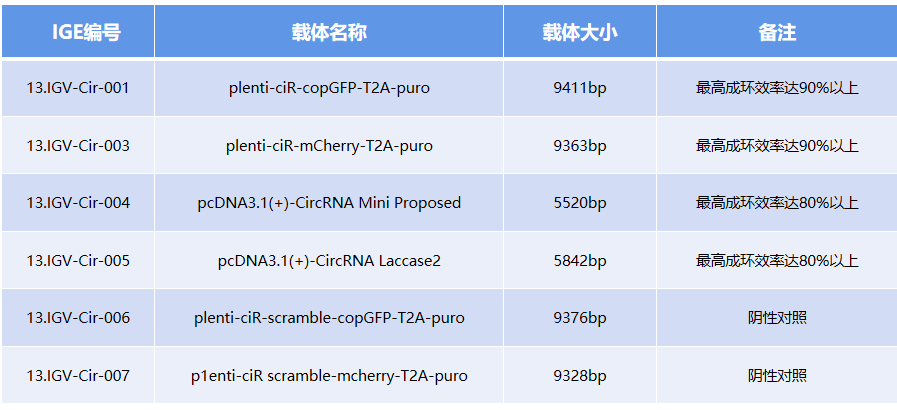
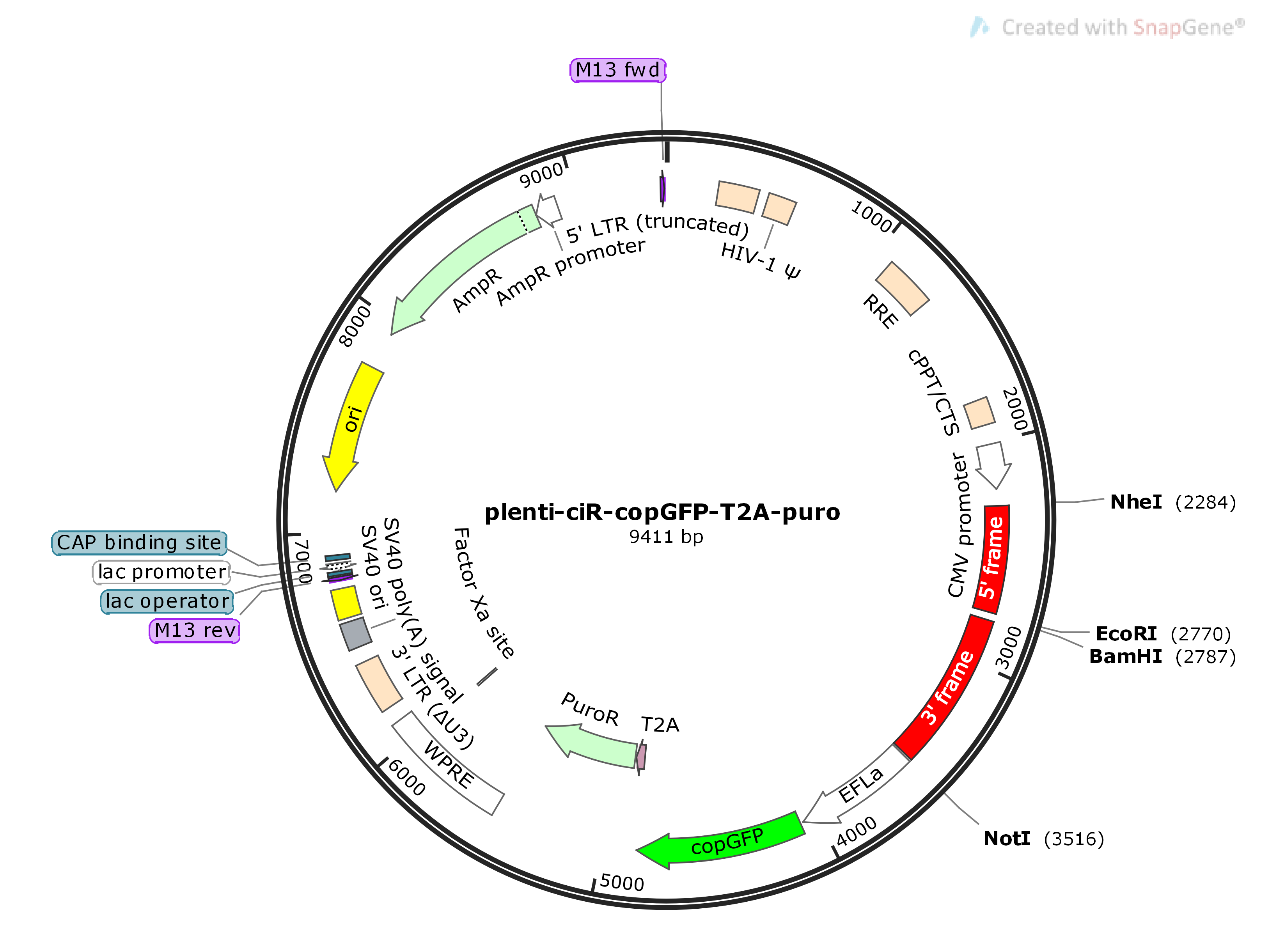
Existing circularization vectors
【1】Wei Dong ,Junming Bi ,Hongwei Liu ,Dong Yan ,Qingqing He ,Qianghua Zhou ,Qiong Wang
Molecular Cancer. 2019 May 17;18(1):95. doi: 10.1186/s12943-019-1025-z.
PMID: 31101108;PMCID: PMC6524247
【2】Yinjie Su ,Weilian Feng ,Juanyi Shi,Luping Chen , Jian Huang , Tianxin Lin .
circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway
Molecular Cancer. 2020 Feb 4;19(1):23. doi: 10.1186/s12943-019-1129-5. PMID: 32019579 PMCID: PMC6998850
【3】Dong Yan,Wei Dong,Qingqing He, Meihua Yang, Lifang Huang, Jianqiu Kong,,Haide Qin.
EBioMedicine. 2019 Oct; 48: 316–331. doi: 10.1016/j.ebiom.2019.08.074.PMCID: PMC6838432;PMID: 31648990
【4】Weikai Xiao,Shaoquan Zheng,Yutian Zou, Anli Yang,Xinhua Xie.
Aging (Albany NY). 2019 Dec 31; 11(24): 12043–12056.doi: 10.18632/aging.102539.PMCID: PMC6949091;
PMID: 31857500
【5】Qingqing He , Lifang Huang ,Dong Yan, Junming Bi , Meihua Yang,Jian Huang , Tianxin Lin.
Aging (Albany NY). 2019 Dec 10;11(23):11314-11328.doi: 10.18632/aging.102530.PMID: 31821171 ;
PMCID: PMC6932899
【6】Haomin Li, Baoli Heng, Peng Ouyang, Xuexia Xie, Tingshun Zhang, Guo Chen, Zheng Chen, Kahong.
Journal of Molecular Histology volume 51, pages317–327 (2020).https://doi.org/10.1007/s10735-020-09882-9

