Long non-coding RNA sequencing service
Service introduction
Long non-coding RNA (lncRNA) is a class of RNA molecules that are longer than 200 nt and do not encode proteins. They regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels by binding to DNA, RNA, or proteins. lncRNA sequencing is used to study all lncRNA and mRNA transcripts that are expressed in a specific tissue or cell type of a species with an available reference genome at a particular time point.
Experimental procedures
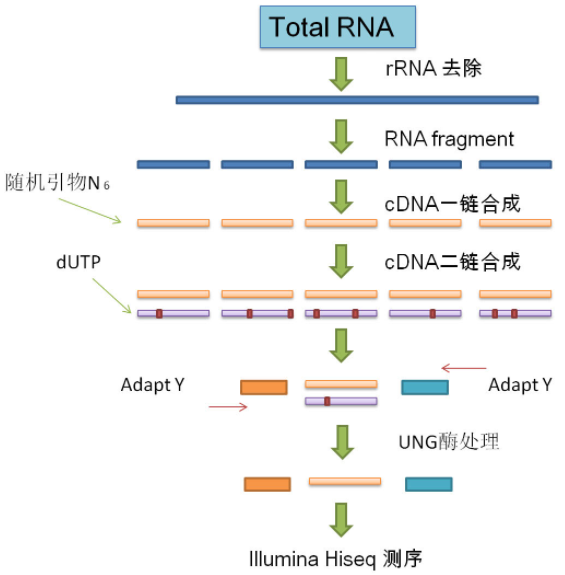
Technical procedures
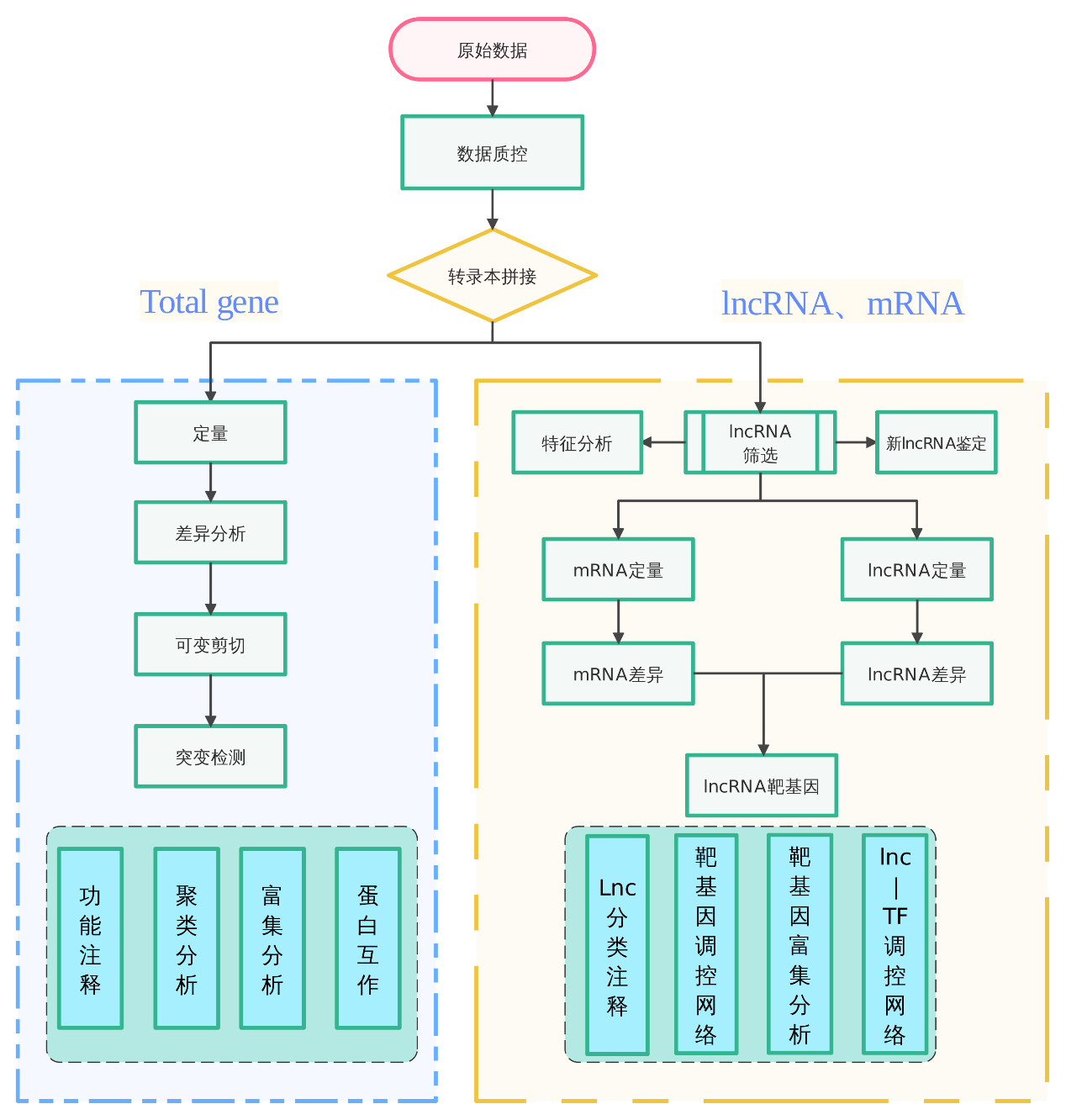
Sample requirements
实验类型 | RNA总量 | 样品浓度和纯度 | 样品质量 | 样品保存 | 样品运输 | 样品选择 |
lncRNA测序 | ≥ 3.0μg | >50 ng/μl,OD260/280 介于 1.8-2.0 之间,无肉眼可见污染 ; | 使用 Agilent Bioanalyzer 仪器检测,则获得的总的 RNA 28S:18S >1,RIN>7.0 | 请选择DEPC 水保存样品,并在样品信息单中注明 | 样品请置于 1.5 ml 管中,并使用封口膜封好,放置足够的干冰运输 | 对于细菌和真菌样品,希望尽量来自于同一个菌株,不要有宿主 DNA 污染 |
Turnaround time
The turnaround time for this service is 45 business days. The sequencing is performed on an Illumina Novaseq6000 platform using paired-end reads of 150 bp. The total raw data output is 12 Gb.
Results overview
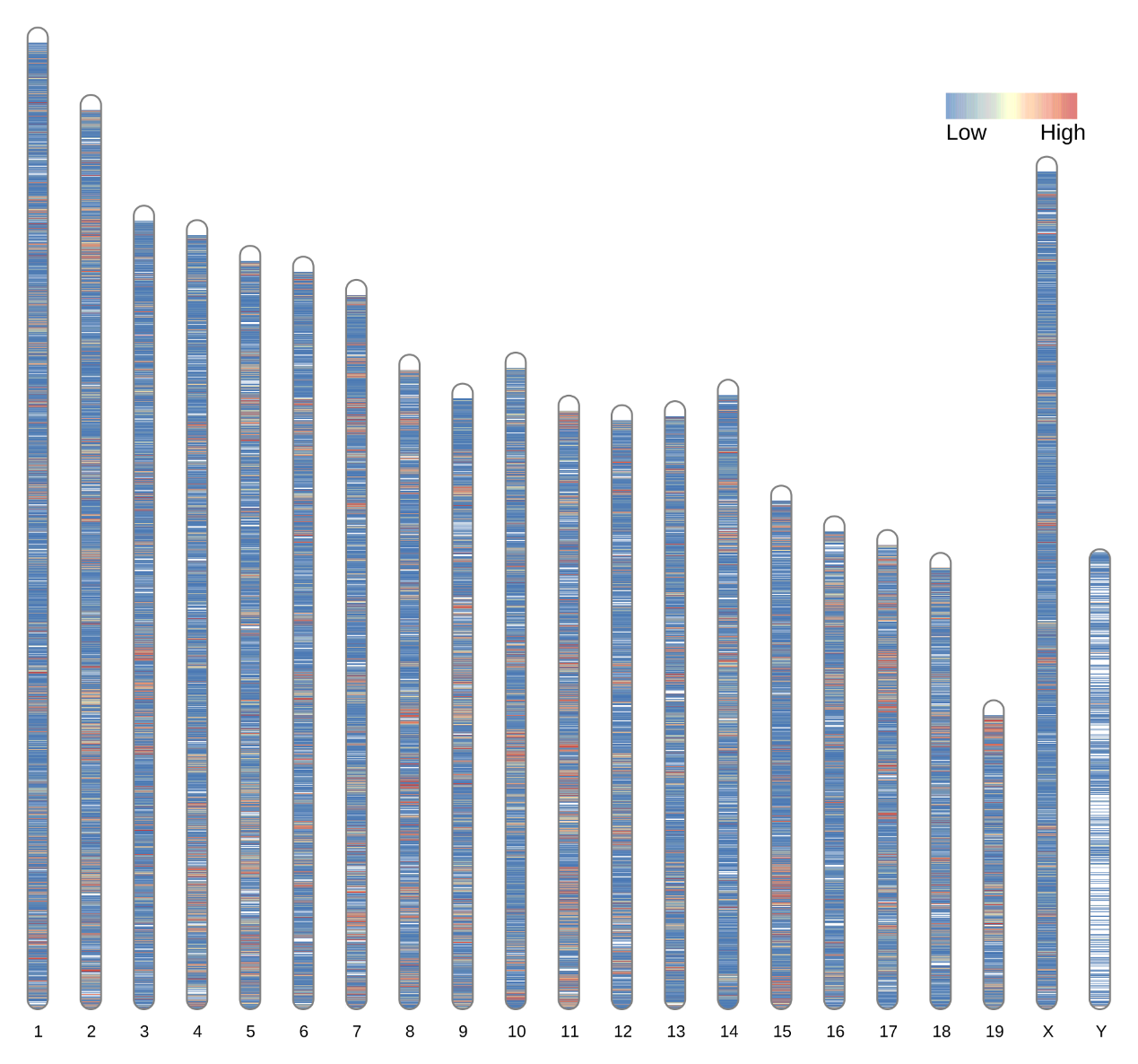
Genome-wide coverage distribution plot: This plot shows the average depth of coverage for each chromosome (scaffold).
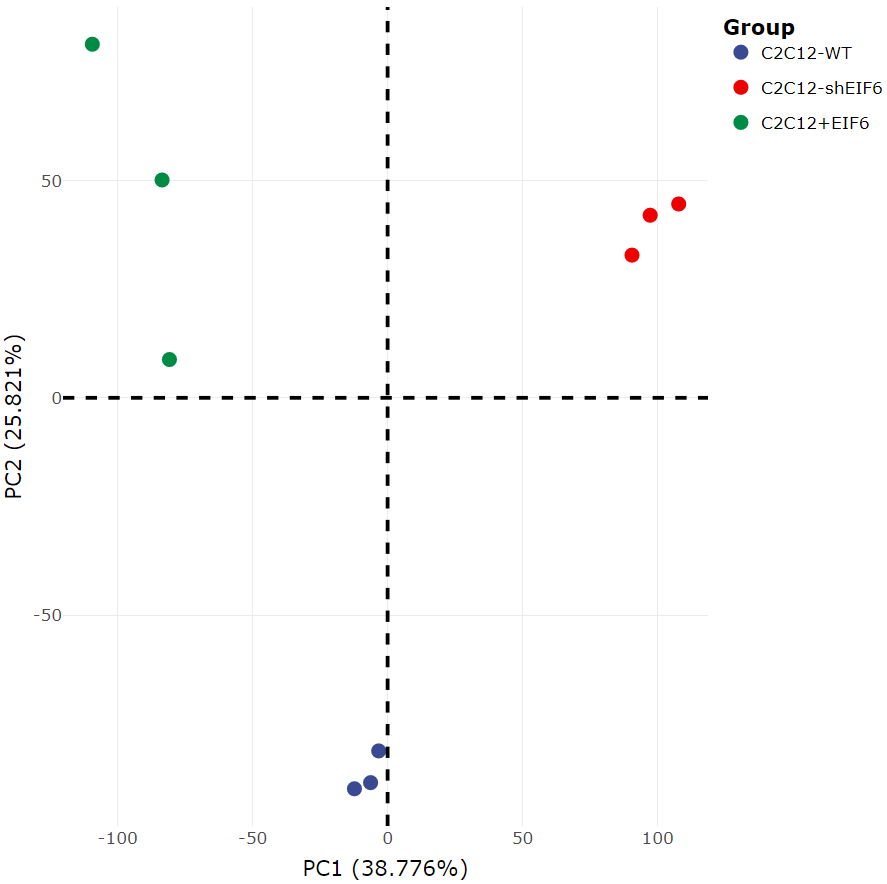
Sample PCA plot: This plot shows the distance between samples based on their read distribution. Samples that are far apart have significantly different read distributions, while samples that are close together have similar read distributions. PCA analysis is often used to assess the reproducibility of samples.

Differential gene volcano plot: This plot shows the expression levels of all genes in the sample, with significant differentially expressed genes highlighted.
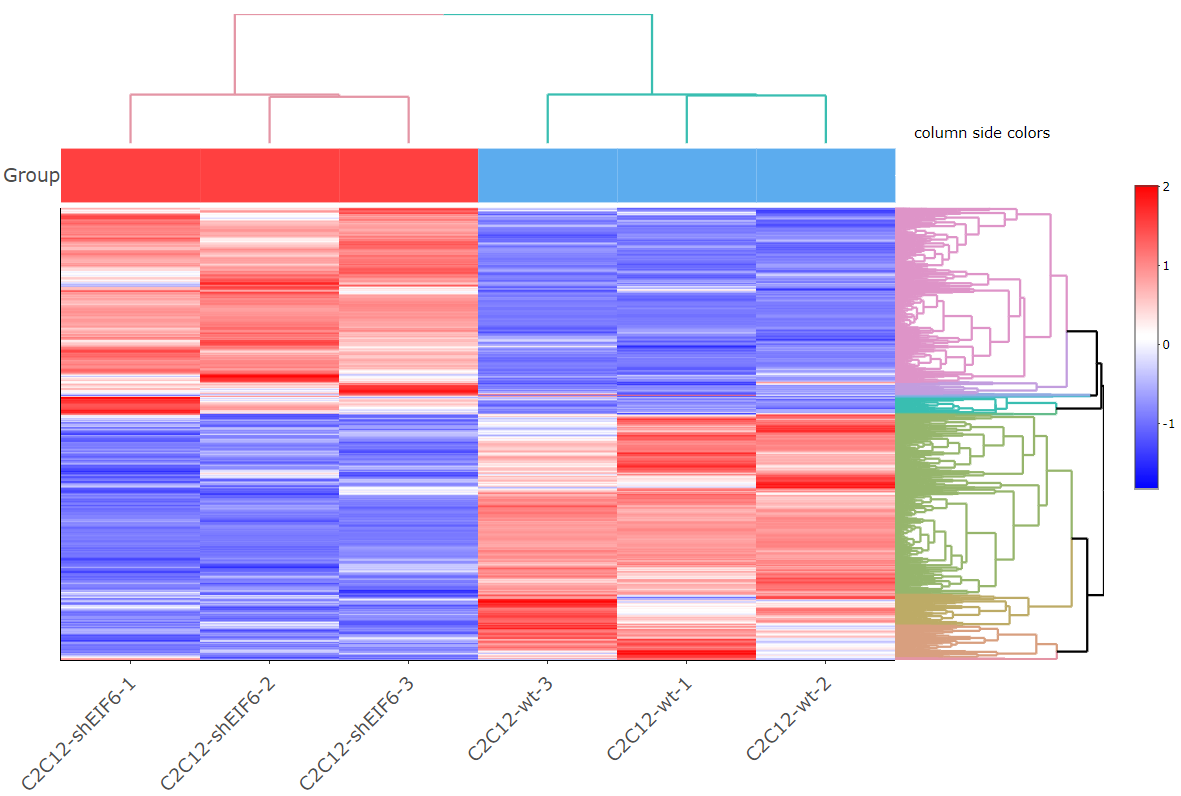
Differential gene clustering heatmap: This plot shows the expression levels of genes in different clusters.
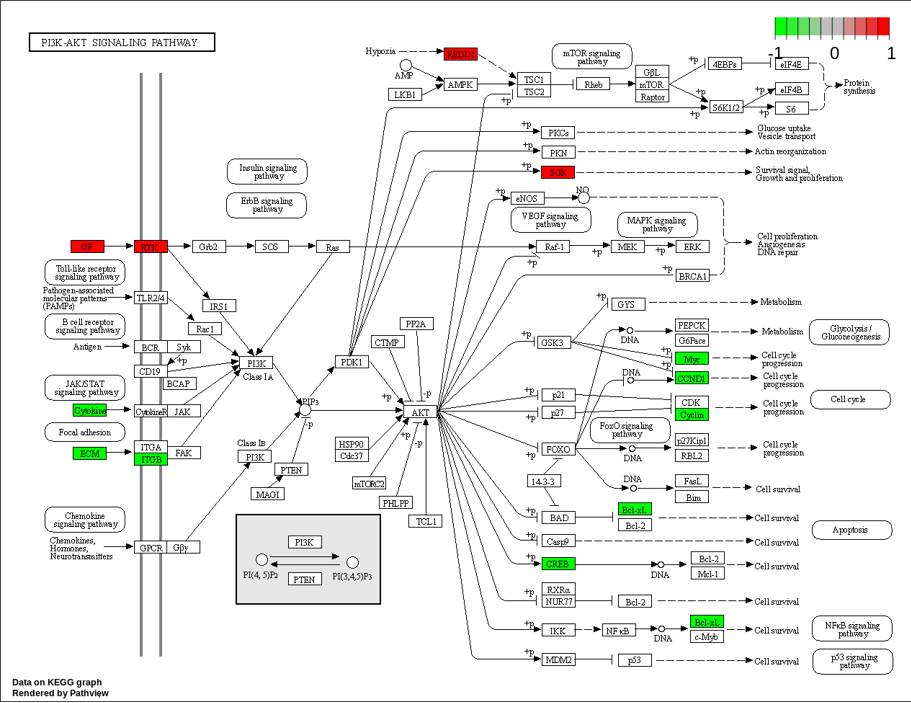
Differential gene KEGG pathway enrichment plot: This plot shows the enrichment of KEGG pathways in differentially expressed genes.

