Restriction-site associated DNA sequencing (RAD-seq) is a high-throughput DNA sequencing method that uses restriction enzymes to digest genomic DNA into fragments that are then sequenced. This approach has several advantages over traditional methods of genome sequencing, including:
Reduced complexity: RAD-seq can significantly reduce the complexity of the genome by targeting only fragments that contain restriction enzyme recognition sites. This can reduce the cost and time of library construction and sequencing.
No reference genome required: RAD-seq does not require a reference genome, which can expand the range of species that can be studied.
High-density SNP detection: RAD-seq can be used to rapidly identify high-density SNP markers, which can be used for genetic variation analysis and population evolution studies.
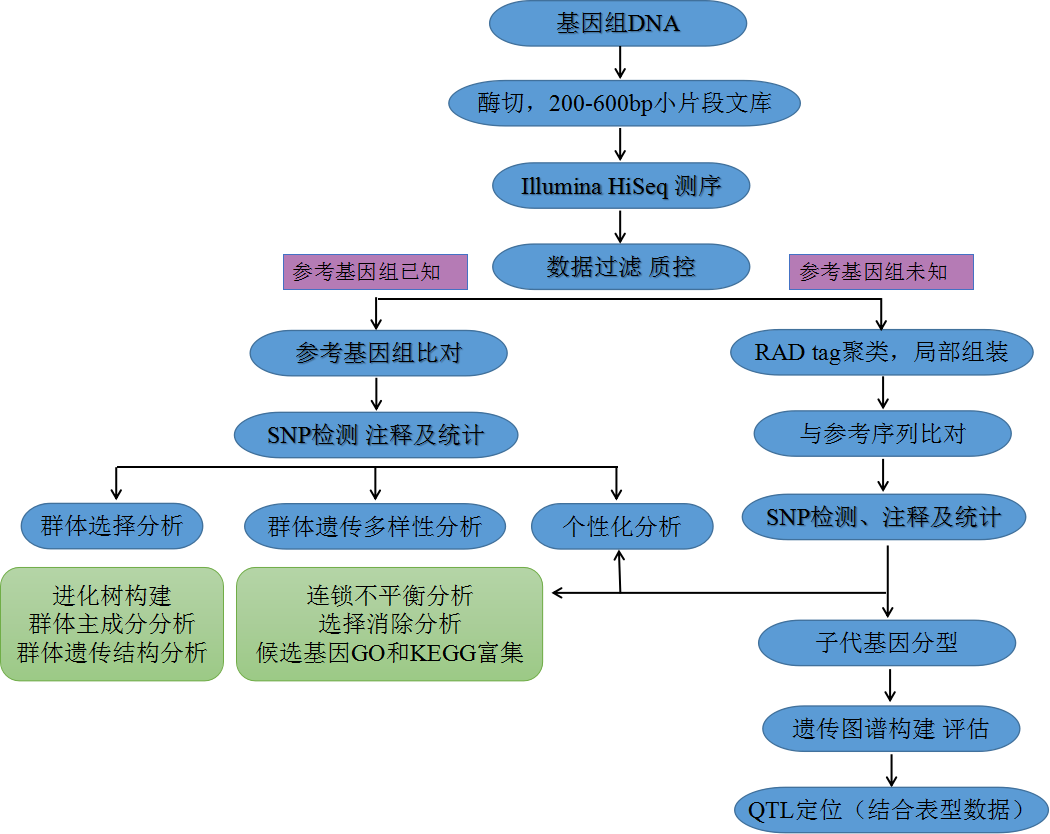
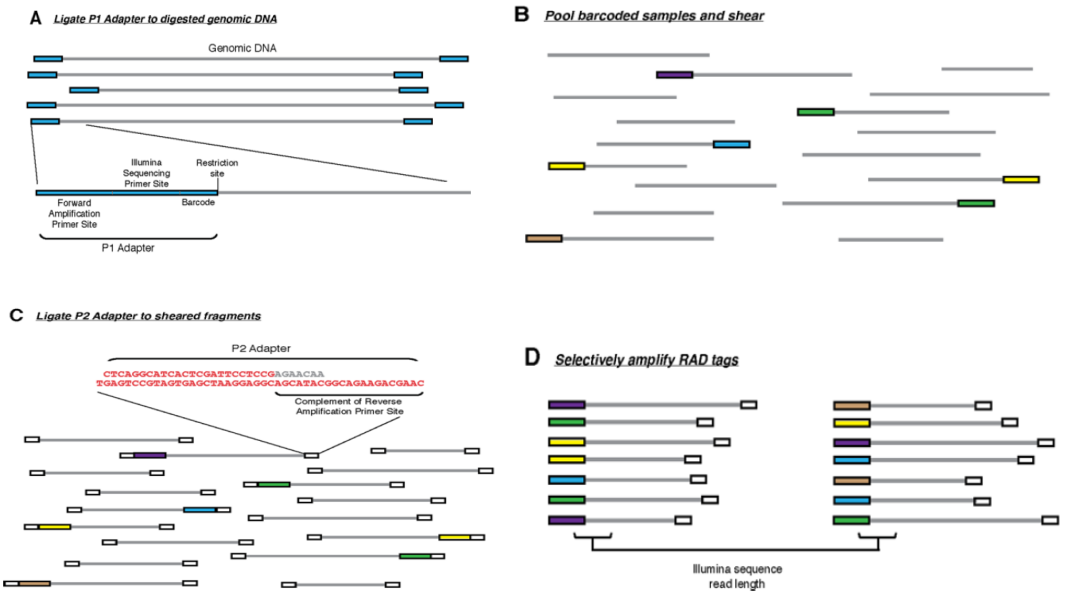

一、Mutation detection:
DNA sample size: ≥ 3 ug
Restriction enzyme: EcoRI (GAATTC)
Recommended sequencing depth: ≥ 1X/sample (assembly sample's sequencing depth ≥ 5X)
二、Genetic map construction
DNA sample size: ≥ 3 ug
Sequencing depth: Parents: 2-5X; Offspring: 0.8X/individual (for temporary populations such as F1 or F2); 0.6X/individual (for permanent populations such as RIL or DH)
Scope of application: Haploid or diploid species; all mapping populations (F1, F2, RIL, DH, etc.); population size: ≥ 100
三、Population evolution:
DNA sample size: ≥ 3 ug
Sequencing depth: ≥ 1X/individual (assembly sample's sequencing depth 5X)
Scope of application: Different subgroups of haploid or diploid species, with clear distinctions between subgroups; each subgroup: 10 individuals or so (≥ 10 for animals, ≥ 15 for plants); total: ≥ 30
四、Sample details
DNA sample:
Concentration: > 100 ng/μl
Total quantity: > 3 μg
OD 260/280: 1.8-2.0
DNA should be free of degradation, with no obvious RNA bands on gel electrophoresis. The genomic band should be clear and complete, with a main band of > 100 kb. If the sample contains polysaccharides or glycoproteins, it will be difficult to break down the DNA, so it is important to ensure that the sample is free of polysaccharides or glycoproteins.
Plant samples:
Young plant tissues should be selected. Each sample should weigh > 500 mg and be shipped frozen on dry ice or liquid nitrogen.
Animal samples:
Fresh animal tissues should be selected, avoiding fatty tissues. Each sample should weigh > 50 mg. For general species, tissues such as liver, kidney, and blood should be selected for sampling. For precious species, tissues such as ear samples or hair (with roots) with low fat content should be used for sampling. To reduce the impact of individual differences on subsequent assembly, samples should be collected from the same individual as much as possible. If the size of the species is small and the amount of DNA extracted from one individual cannot meet the requirements of the sequencing experiment, the number of sampled individuals should be reduced as much as possible while ensuring the amount. The tissue sample should be > 50 mg, and as much as possible should be provided, as the DNA yield of different species varies.
Sample transportation:
The sample tube must be clearly labeled with the sample number. The tube opening should be sealed with Parafilm membrane. The sample should be shipped frozen on dry ice or ice packs. Dry ice transportation time should not exceed 72 hours; ice pack transportation time should preferably not exceed 24 hours. The sample should be avoided from repeated freezing and thawing during storage.
1. Q:What factors should be considered when developing high-density SNPs markers?
A:The basic conditions and design concept of high-density SNPs marker development is that the fragments obtained are evenly distributed on the genome as much as possible, and thousands of SNPs markers are obtained by measuring a small number of sequences that represent the whole genome information. First, we need to use bioinformatics methods to systematically analyze the reference genome (or known BAC sequence) of the target species, and according to the genome GC content, repetitive sequence situation and gene characteristics, select the corresponding restriction enzyme and sequencing library type to ensure that the density, uniformity and efficiency of molecular markers meet the needs of genetic analysis and molecular breeding.
2. Q:What is the scope of application of RAD-seq?
A:It can be used to identify the genetic map, polymorphism (phylogenetic analysis), association, QTL location, etc. of any species (with or without reference genome data). RAD-Seq technology can be widely used in mutation detection, genetic map construction, functional gene mining, population evolution, etc.
3. Q:What are the advantages of using RAD-seq to study population evolution?
A:Generally speaking, population evolution research involves a large sample size. RAD-seq can significantly reduce the complexity of the genome, reduce the cost of library construction and sequencing, and make the operation simple. In addition, it is not limited by the reference genome, and the research range of species is wider. RAD-Seq is especially suitable for large sample size research, and can lay a solid foundation for in-depth information mining using whole genome resequencing technology.
4. Q:What is the difference between with-reference and without-reference?
A:For the discovery of mutation information, the efficiency and accuracy of sequence alignment and mutation detection are higher with reference genome. It can also locate genes under selection and study the phenomenon of linkage disequilibrium in the population.

