Service Introduction
IGEBio has a number of ABI 3730XL sequencers and has over ten years of experience in Sanger sequencing. We have developed a series of solutions for samples of varying complexity, and we have expanded the applications of our Sanger sequencing platform to multiple upstream fields of molecular biology, including:
SNP detection
Fragment analysis
SSR/STR typing
Gene detection
Strain identification
Mitochondrial sequencing
Genome sequencing
Gel running
Download
Service Advantages
Stable quality: Accurate read length of over 800 bp, effective read length of over 900 bp.
High accuracy: QV20+ and CRL value >1000.
Strong timeliness: Daily processing capacity >2000 reactions, timely and accurate results, with results
available in as fast as 24 hours.
Challenging templates: Mature solutions for high GC, AT rich, hairpin structure, and repetitive sequences.
One-stop service: Genome extraction, primer design, PCR amplification, Primer Walking, sequencing, and
result analysis.
Technical Procedures
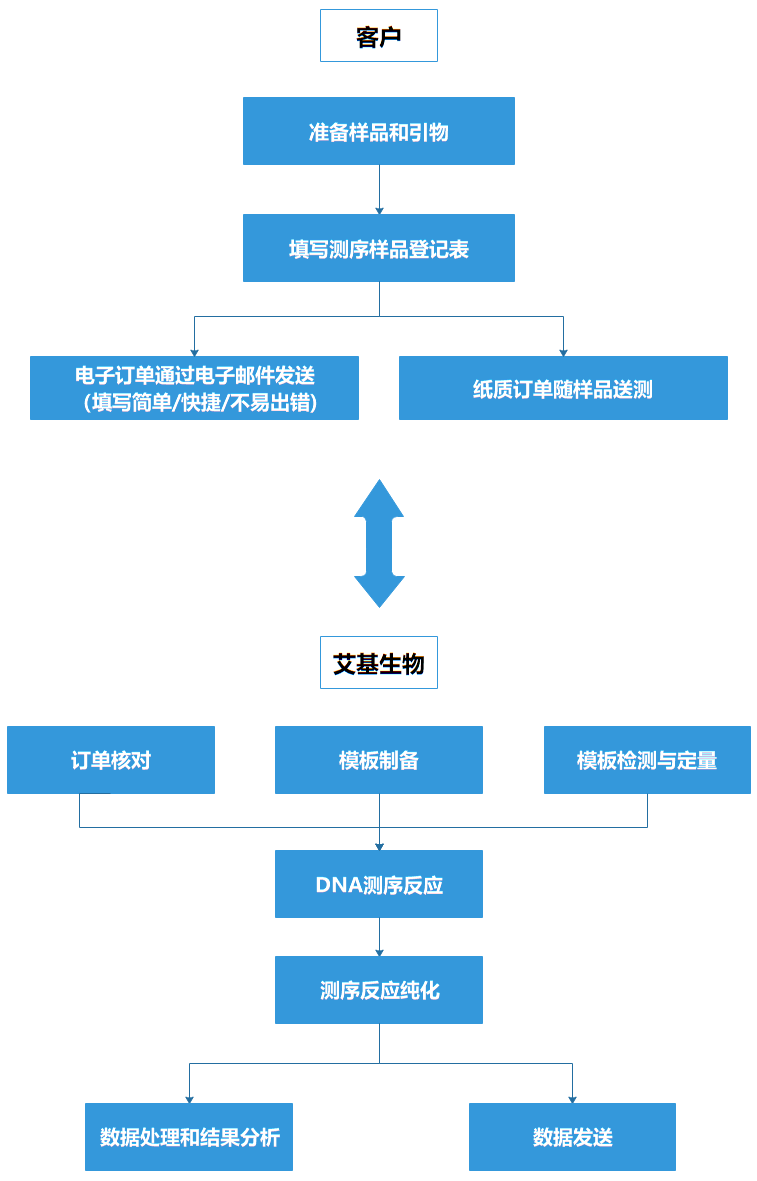
Sample Requirenments
【Cultures and plates】
The culture needs to be overnight incubated, at least 200 ul is required. For samples that do not need to be
cultured and can be extracted directly, please provide 2-4 ml of overnight incubated culture.
Please indicate the antibiotic resistance of the sample. Common antibiotics include ampicillin (Amp),
kanamycin (Kan), and chloramphenicol (Chl).
If the plasmid contained in the culture is of ultra-low copy number, please provide the plasmid directly.
Except for E. coli, please provide the plasmid directly for cultures of other bacteria.
【Plasmids】
Concentration >50 ng/ul, total volume not less than 10 ul.
Please ensure that the plasmid is dissolved in sterile double-distilled water. Plasmids dissolved in TE will
cause the reaction to fail.
【PCR products】
Purified PCR products: Please ensure that the DNA is dissolved in sterile double-distilled water, concentration
>30 ng/ul, total volume not less than 20 ul.
Unpurified PCR products: Please indicate the fragment length, whether the band is single, and the original
liquid volume is not less than 20 ul.
Fragments <100 bp are recommended to be cloned for sequencing, and fragments >4 kb are recommended
to be sequenced in segments.
【Sequencing primers】
Concentration ≥5 pmol/ul, total volume not less than 10 ul, and ensure that it is dissolved in sterile double-
distilled water.
The length of the sequencing primer should be controlled as much as possible within 20-30 bp. Primers that
are too short or too long will affect the quality of the sequencing.
Primers containing merged base fluorescent tags or modifications are not suitable for sequencing.
Notes
The sample registration form must be filled out in detail according to the requirements to avoid
experimental delays caused by problem orders.
Ensure that the sample and the registration form name are consistent. It can contain "letters", "numbers", and
"-". Avoid the appearance of Chinese characters, Latin letters, and other special characters.
Samples marked "sequence through" will be spliced by default. For samples marked "double-directional"
sequencing, please indicate if splicing is required.
If the sample is a culture or plate, please indicate the type of antibiotic.
If the sequencing primer is not a general primer, please provide it yourself or entrust it to be synthesized.
Select the primer type (general/new/sent/need to be synthesized) and indicate the concentration.
If the plasmid sent for measurement is not a common vector, the general primer should be written with the
full name of the primer.
If the unpurified PCR product has already been detected as a single band, please indicate it in the order,
which can greatly improve the success rate of sequencing.
For samples with special structures, such as: High GC, AT rich, repetitive sequences, hairpin structures, please
note on the order.
For a large number of samples, please use an electronic order, which is convenient for verification and less
likely to make mistakes.

