Service introduction
siRNA, or small interfering RNA, is a short double-stranded RNA molecule that can be used to silence gene expression in cells. siRNAs are typically 19-23 nucleotides long and are designed to target a specific gene sequence. Once inside the cell, siRNAs are cleaved by an enzyme called Dicer, which releases the two strands of the RNA. One strand, called the guide strand, then binds to the target mRNA molecule, causing it to be degraded. This prevents the mRNA from being translated into protein, resulting in the silencing of the gene.
SiRNA synthesis is a simple and efficient way to achieve gene silencing in cells. It is a low-cost, high-throughput method that is also well-suited for clinical applications.
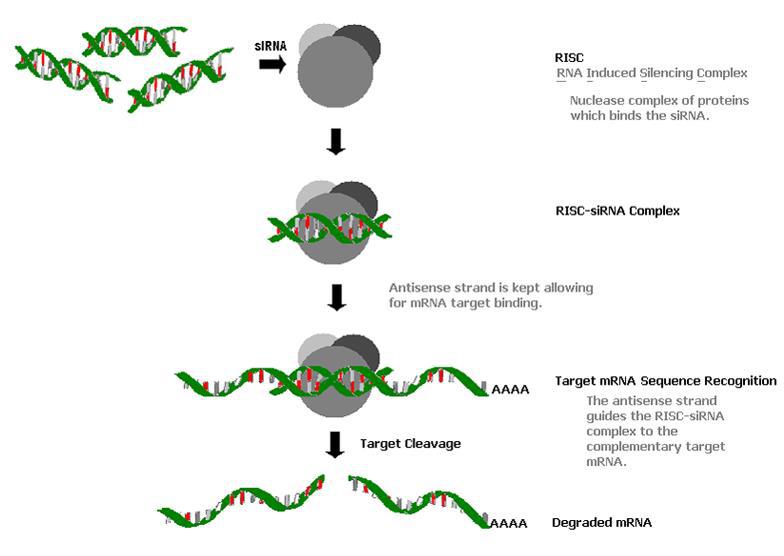
Gene Silencing Mechanism
Types of siRNAs
Conventional siRNAs are chemically synthesized and are designed to target a specific gene sequence. They are available for a wide range of genes, including human, mouse, and rat.
Control siRNAs
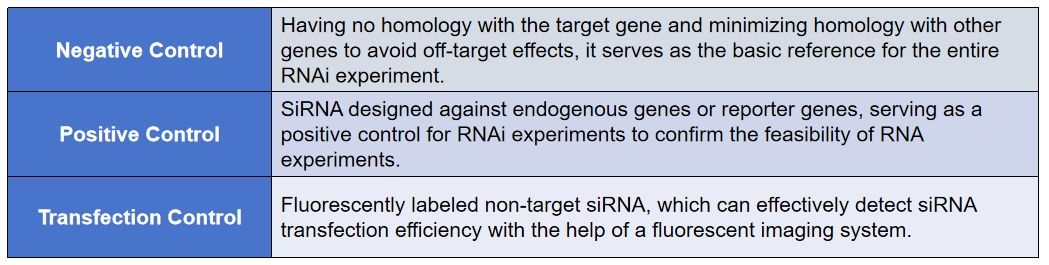
Control siRNAs are used to validate RNAi experiments. They include transfection controls, positive controls, negative controls, and blank controls.
Transfection controls
Transfection controls are used to determine the efficiency of siRNA transfection. They are typically labeled with a fluorescent dye, such as Cy3, which allows for easy visualization under a fluorescence microscope.
Positive controls
Positive controls are used to confirm that the siRNA is targeting the correct gene. They are typically designed to target a gene that is known to be expressed in the cell type of interest.
Negative controls
Negative controls are used to ensure that the siRNA is not targeting any other genes. They are typically designed to target a gene that is not expressed in the cell type of interest.
Chemically modified siRNAs
Chemically modified siRNAs have been developed to improve their properties, such as stability, solubility, and delivery. Common modifications include:
2'-O-methyl modifications: These modifications reduce the susceptibility of siRNAs to degradation by nucleases.
2'-O-fluoro modifications: These modifications also reduce the susceptibility of siRNAs to degradation by nucleases.
Cytosine modifications: These modifications can improve the solubility of siRNAs.
Biotinylation: This modification allows siRNAs to be labeled with biotin, which can be used for detection or purification.
Download
IGE-Routine siRNA Synthesis Order

